r_eda
占比选图
参考 Strayer (2019)
对于占比的表示,一共有三种图表可以展示
- 圆饼状图,pie,用角度度量
- 华夫饼状图,waffle,用面积度量
- 累计直方图,用部分面积度量,取极坐标就是圆饼状图(参考相关 ggplot2 的代码)
当数据的类别超过3个时,圆饼状图很难进行比较。
Intuitively, you can think about a pie chart as a stacked bar chart that has been ‘wrapped’ around some central axis. DataCamp
pie
圆饼图和累计直方图类似。 在 ggplot2 中只需要两个函数
coord_polar(theta = 'y')x = 1
suppressMessages(library(tidyverse))
who_disease <- read_csv('datasets/who_disease.csv')
## Parsed with column specification:
## cols(
## region = col_character(),
## countryCode = col_character(),
## country = col_character(),
## disease = col_character(),
## year = col_double(),
## cases = col_double()
## )
# Wrangle data into form we want.
disease_counts <- who_disease %>%
mutate(disease = ifelse(disease %in% c('measles', 'mumps'), disease, 'other')) %>%
group_by(disease) %>%
summarise(total_cases = sum(cases))
pie_raw <-
ggplot(disease_counts, aes(x = 1, y = total_cases, fill = disease)) +
# Use a column geometry.
geom_col() +
# Change coordinate system to polar and set theta to 'y'.
coord_polar(theta = 'y')
pie_raw
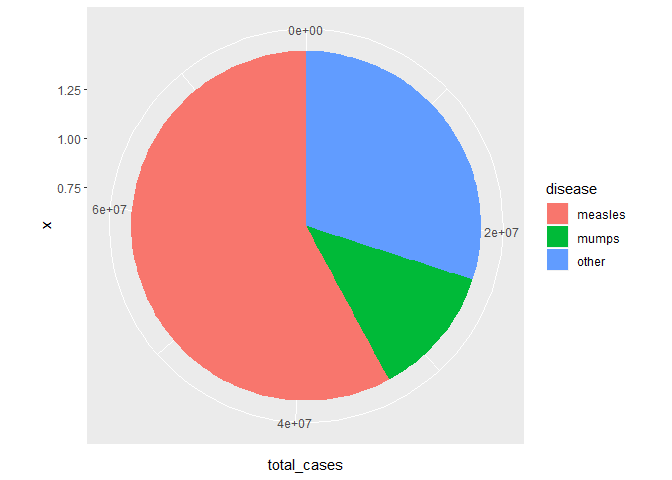
可见 pie 不美观,设计中间变量 pie_raw 进行美化。
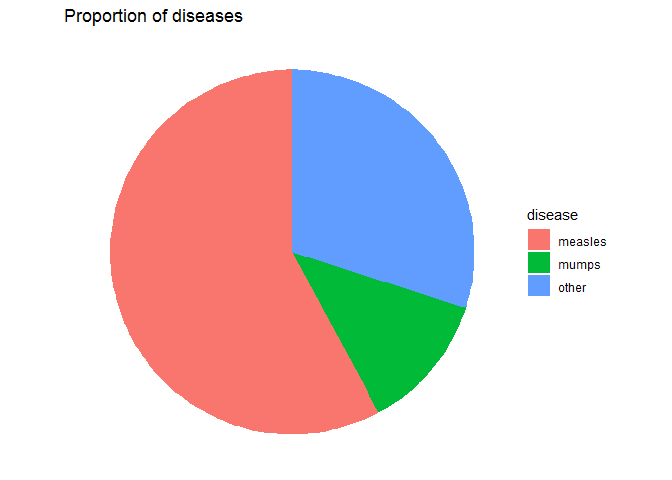
pie_raw +
theme_void() +
# 做一定清洗
labs(title = 'Proportion of diseases')
waffle
由上可知,当 class 变多,pie 就不适用了。
disease_counts <- who_disease %>%
group_by(disease) %>%
summarise(total_cases = sum(cases)) %>%
mutate(percent = round(total_cases/sum(total_cases)*100))
# Create an array of rounded percentages for diseases.
case_counts <- disease_counts$percent
# Name the percentage array with disease_counts$disease
names(case_counts) <- disease_counts$disease
# Pass case_counts vector to the waffle function to plot
library(waffle)
waffle(case_counts)
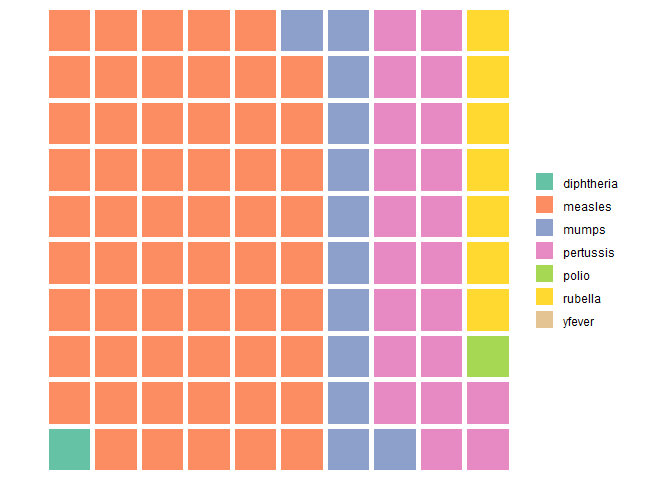
注意这里 waffle 的 input 是
parts named vector of values to use for the chart
是一个已经命名的向量。
stacked bar
当比例除了需要组内比较,还需要组间比较,就需要累计直方图了。
disease_counts <- who_disease %>%
mutate(
disease = ifelse(disease %in% c('measles', 'mumps'), disease, 'other') %>%
factor(levels = c('measles', 'other', 'mumps')) # change factor levels to desired ordering
) %>%
group_by(disease, year) %>%
summarise(total_cases = sum(cases))
# plot
ggplot(disease_counts, aes(x = year, y = total_cases, fill = disease)) +
geom_col(position = 'fill')
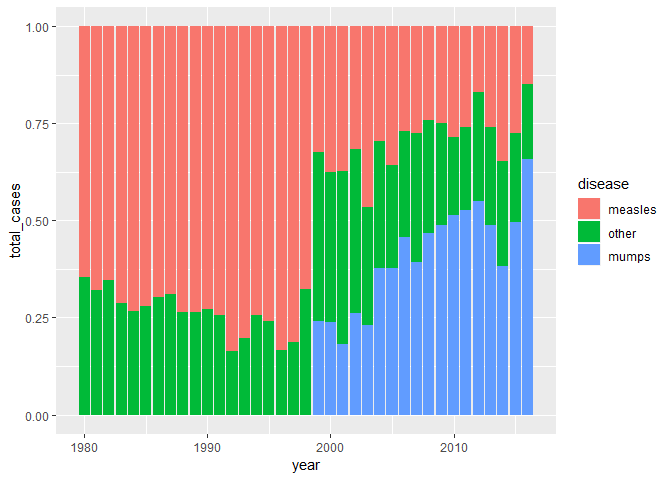
当调整了分类变量的 levels 后,可以主要观察 measles 和 munmps 的占比组间变化了。
Just remember to keep the number of classes low to aid readibility! DataCamp
因此这也暴露了一个问题,bar 和 pie 都不能容纳太多的 levels。
Strayer, Nick. 2019. “Visualization Best Practices in R.” DataCamp.
2019.
<https://www.datacamp.com/courses/visualization-best-practices-in-r>.